Aberration Scoring
Digital Helpers for the Assessment of Chromosomal Aberrations
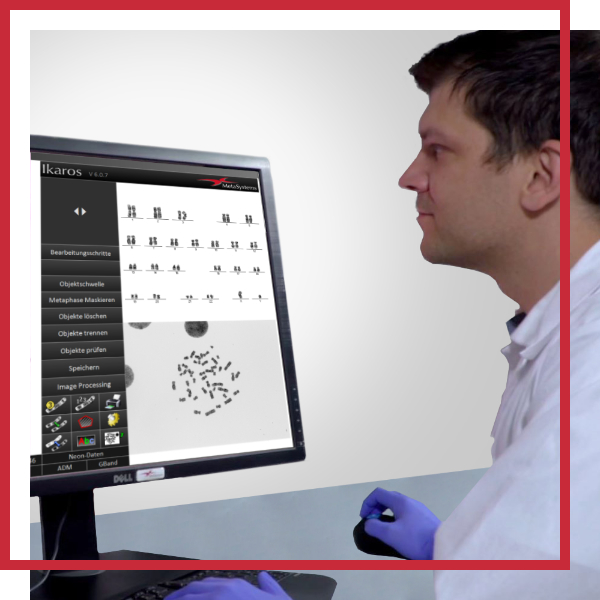
You may benefit most from the following customizations:
- A user-friendly and comprehensive software environment for establishing a digital workflow to assess structural chromosomal aberrations.
- Effortless integration of a routine for automated metaphase detection and image acquisition.
- Comprehensive documentation of all images, processes, and results facilitates the configuration of a workflow following GLP standards.
- The option to configure automatic detection of dicentric chromosomes according to user-defined parameters.
- Images and/or original metaphases can be analyzed on-screen, with aberration counts entered into customizable scoring sheets.
- A flexible report generation option that provides access to all entered aberration and study data.
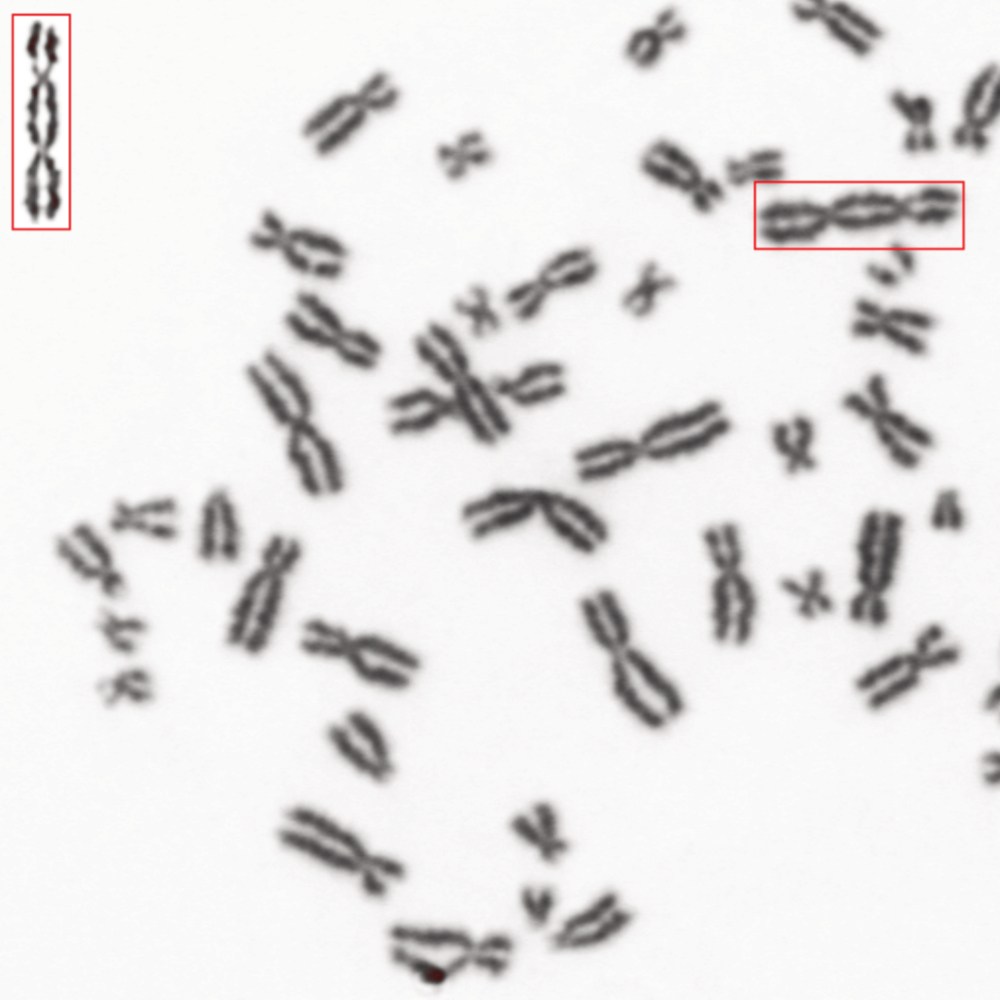
The quantification of chromosomal aberrations is commonly considered the gold standard for detecting DNA damage in both biological dosimetry and genetic toxicology testing. However, unlike other tests, aberration analysis under the microscope is complex and time-consuming.
Using Metafer's image acquisition, processing, and management tools, our application specialists have elaborated routines for many customers that assist in aberration evaluation and digitize much of the workload. These workflows combine the possibilities that Metafer offers for detecting objects, in these cases, metaphases, and for image acquisition, with the configuration of work environments that allow the images to be easily analyzed on the screen. Implementing user-configurable scoring sheets enables the aberration data to be entered with a few simple mouse clicks or keystrokes and saved alongside the images. Once recorded, the data can be later generated into customizable reports. These reports can include additional information on the specific study and be printed or exported as a data file (e.g., for import into another database). An integrated, interactive Chi-square calculator enables rapid statistical evaluation of the recorded data.
For users seeking further simplification in chromosome aberration analysis, automated detection of dicentric chromosomes (DICs) can be implemented. DICs, which are chromosomes with two centromeres resulting from the rearrangement of two damaged chromosomes, are commonly used as key markers in chromosomal aberration studies. With the workflow, implemented and validated by numerous laboratories, DICs are automatically identified and counted in a high-throughput, unattended mode. All detected DICs are highlighted in the metaphase, and the corresponding scoring sheet displays the count of dicentric chromosomes in the selected metaphase, with the option for manual adjustments. During review, the metaphase can be easily centered under the microscope for further examination. The scoring parameters are user definable and can be adjusted with an integrated training module.
Our application specialists can collaborate with you to establish a routine using the Metafer Platform Software, enabling the identification and capture of metaphases from your specimens as high-resolution images. More information on this option can be found here. If metaphase images are available, a workspace can be set up within the software to display the images alongside a scoring sheet. You can then assess the metaphase on the screen and input the corresponding aberration data into the scoring sheet, either by clicking the mouse or using the keyboard. The data will be saved along with the images and can be compiled into customized reports. Various tools are available for generating these reports, and you can choose to print them, save them as PDFs, or export the raw data to an external database. If you are interested in automatic dicentric chromosome recognition, we can customize a parameter file, which will be validated by you. The results will be shown on the screen, allowing you to confirm or adjust them in the scoring sheet.
Yes, we have developed parameter sets for many other customers that identify dicentric chromosomes in similar preparations. Our application specialists will be happy to design a tailored workflow for you, which you can implement and validate on-site.
The Metafer Platform Software can be seamlessly scaled with the addition of suitable hardware components. This allows up to 800 specimens to be scanned unattended in a single session, including overnight or during weekends if needed. Features such as automated barcode label capture and the automatic application of immersion oil for acquiring high-resolution images can also be configured.
MetaSystems software provides, among other functions, features to assist users with image processing. These include, but are not limited to, the use of machine and deep learning algorithms for pattern recognition. The output generated in this process should be regarded as preliminary suggestions and, in any case, mandatorily requires review and assessment by trained experts.
MetaSystems offers Customization Packages for application workflows that have been successfully implemented for customer labs using standard Metafer platform functionality. It is expected that they can be implemented for other customer labs using similar workflows and slide preparation procedures. If a Customization Package is purchased, MetaSystems product specialists will – based on their experience from other similar application cases - support the customer lab in adapting the Metafer software configuration to their needs. The performance of the solution will depend on the quality of the customer slides and the expertise of the users, MetaSystems cannot specify or guarantee any performance parameters. The validation of the solution for clinical use is the sole responsibility of the customer lab.














